Protein molecules are the building blocks of life
All living things that we know of use protein molecules for structure and function. Nearly everything in your body is made of various kinds of proteins. Special proteins called enzymes catalyze nearly every chemical reaction that occurs in biological systems.
A fundamental goal of biological science is to understand every protein in terms of its structure and function. The 3-dimensional structure of a given protein — its 3D shape — is crucial to its function.
Proteins are polymers, long chains of linked smaller molecules, 20 types of amino acids.
polymer
In its Greek origins, poly means many and meros means shares or things — "many things". In chemistry, a polymer is a long chain of linked molecules of the same or similar type. Polyethylene is a polymer of ethylene molecules, often thousands of units long. Proteins and DNA are biological polymers of amino acids and nucleic acids, respectively.
Amino acids
Amino acids are small organic molecules that contain a central carbon atom, called the alpha carbon, or Cα, bonded to a hydrogen (H) atom, an amine group (-NH2), a carboxylic acid group (COOH), and a so-called side chain, abbreviated R in the diagram below.
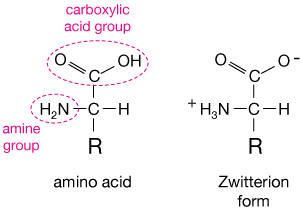
All amino acids have the first three of these elements, but the 20 naturally-occuring amino acids all have a different side chain. The simplest side change, on the amino acid alanine, is simply a hydrogen at the position of the R. Other side chains are more complicated.
If you know your acid and base chemistry (don't worry, you can understand this section pretty well if you don't), you'll know that the proton (H+) on the OH arm of the carboxylic acid group is the one that ionizes in water, and that the NH2 (amine) group, just like ammonia, can accept a free proton from solution, and thus acts as a base.
In fact, in water solution near neutral pH (pH = 7), we always find what is called the zwitterion form of an amino acid, shown on the right in the figure.
Proteins are long chains of amino acids linked through their amino and carboxylic acid ends. The backbone of the chain is the same in any protein, sequence of the side chains, and how they interact with other nearby side chains (and water molecules) is ultimately what gives any protein its function.
Let's take a look at the different kinds of amino acids, grouped according to some general properties.
Aliphatic amino acids
The alaphatic amino acids all have, not surprisingly, aliphatic side chains. Aliphatic means "fat- or oil-like." Notice that these side chains, except for methionine which contains a sulfur (S) atom, only contain carbon and hydrogen. These side chains don't mix well with water — they're like oil in water — and they tend to move toward the inside of a
protein, away from the surrounding water.
Each amino acid has a name (these usually aren't capitalized), a three-letter symbol like gly for glycine or Ile for isoleucine, and also a single-letter code (for efficiency when writing long sequences). Each kind of label is shown in the figures.
Aliphatic side chains are also said to be hydrophobic.
hydrophobic
Literally (from Greek words), hydrophobic means water (hydro) fearing (phobic). Hydrophobic compounds are not soluble in water and are not by wetted by water solutions (think about water beading on a piece of glass). Contrast these to hydrophilic compounds (water loving), which dissolve easily in water.

Polar amino acids
The polar amino acid side chains all contain something that resembles water (like an OH group) or ammonia, like an NH2 group. Cysteine is a special case because it can form a disulfide bond with another cysteine. We'll cover that later. Polar side chains are hydrophilic.
Notice that the amine group of proline is tied to its side chain, the side chain doubling back to form a five-membered ring. That creates a geometric restriction plays a valuable role in determining the 3D structure of a protein containing one or more prolines. We'll come back to it later.

Acidic amino acids
These amino acids have a carboxylic acid side chain (shown ionized here as they would be in approximately neutral solution). These amino acids, along with the basic and polar amino acids, are generally exposed to water in solution; they generally reside on the outside of a protein blob because they are hydrophilic.
There are only two amino acids with acidic side chains, aspartic acid and glutamic acid.

hydrophilic
Literally (from Greek words), hydrophilic means water (hydro) loving (philic). Hydrophilic compounds are soluble in water and can by wetted by water solutions. Contrast these to hydrophobic compounds (water fearing), such as oils, fats and waxes, which do not dissolve well — or often at all — in water.
Basic amino acids
The basic amino acids all have exposed N, NH or NH2 groups (some are shown with an extra proton, as they would have in neutral solution).
There are three amino acids with basic side chains: lysine, arginine and histidine. All of these side chains are hydrophilic.

Aromatic amino acids
The aromatic side chains contain what are called conjugated rings. In the right circumstances, these are called "aromatic" rings. Aromatic compounds have familiar smells, like the smells of bananas and oranges.
There are three amino acids with aromatic side chains: phenylalanine, tyrosine and tryptophan. The latter two absorb UV light at a wavelength near 280 nm, and are useful in estimating the concentrations of proteins using spectroscopy.

The making of a protein
Proteins are composed of tens to (usually) hundreds of amino acids, each linked in a chain by a peptide bond. Peptide bonds are formed by enzymes which have that specific job. The making of a peptide bond is a condensation reaction, because a molecule of water is formed in the process. It looks like this:

In this reaction, the peptide bond is formed as the OH from the left amino acid and the H from the right one detach and combine to form a water (H2O) molecule. The new bond (red line) between the alpha carbon and the amino nitrogen is called a peptide bond, and the di-amino acid formed is called a dipeptide. In just this way, long chains of amino acids are linked to form proteins.
Often when the chain length is less than 30 amino acids or so, the product is called a peptide instead of a protein. As we will see, proteins have properties that small peptides can't have because of their size.
Protein sequences
The figure below might seem a little complicated, but it shows a lot. Focus first on the first line of every block of letters, the one labeled "H CBP." As you read from left to right, top to bottom, this line gives the single-letter code of a small part of the sequence of amino acids in a human protein called "CBP." CBP is an important factor in how genes are turned on and off, although as of this writing, it's still a bit mysterious.
The first few amino acids in the sequence are S, G, N, I, G, which are serine-glycine-asparagine-isoleucine-glycine. You can see why the single-letter code is preferable.
Also shown in the figure are partial sequences of three other proteins, M-CBP is the version found in mice; HP300 is a very similar human protein; and D-CBP is CBP from the fruit fly, Drosophila melanogaster.
Amino acids highlighted in black are identical between these species, and those in gray are amino acids that aren't quite identical,
but are of a similar type and size.
Charts like this, called alignments, tell us a lot about the evolution of species and about which parts of proteins do important jobs that are common between species. For nature to have preserved this sequence among species as diverse as human, mouse and fruit fly, it must have some basic, important purpose.
There are other sections of these proteins that aren't nearly so similar; there natural selection has produced enough changes over time to make the sequences very different.
Notice also that the fruit fly protein has a 29-AA insert that the other species don't have. It's not known why that's there. We know from some other examples that sometimes a "loop" of extra amino acids just hangs off the protein not doing anything, but other examples tell us that such an insert performs some other job in the other species.
Alignment of CBP protein segment across species

Residues
Often, an individual amino acid in a protein chain is referred to as a "residue." One might refer to "the second alanine residue," or "residues 50-100", for example.
Four levels of structure
1. Primary (1˚) structure
The simple sequence of amino acids linked in a chain, like a string of amino acid "pearls," on the right, is the first of what we call "levels of structure" of a protein. The sequence is the primary (1˚) structure. Because there are 20 choices of amino acid, there is practically an infinite number of sequences.
Think about a simple peptide of 10 amino acids: There are 20 choices for the first residue, and 20 for the second. That's already 20 x 20 = 400 different combinations. There are 2010 = 10,240,000,000,000 possible 10-AA peptides (10 trillion!), and most proteins contain hundreds of amino acids. You can see why there is an almost infinite variety of protein molecules. Who knows what nature hasn't invented yet?
The sequence of a protein is the foundation for what is to come. If we change the sequence, then all of the other structural features may (or may not) change.

2. Secondary (2˚) structure
The secondary structure (2˚ structure) of a protein comes from interactions between side chains. Go back and take a look at some of the side chains in the charts above. They can form weak bonds with one another in many ways.
Much like the hydrogen bonds that form in water, H-bonds can also form between polar amino acids. Acidic, basic and aromatic side chains can also link up with weak bonds. By weak I mean much weaker than a peptide (covalent) bond, but still strong enough to cause some stable structure to develop along the chain.
Without any significantly polar or charged side chains, a string of amino acids won't develop any long-lasting structure. For example, a long string of glycine molecules, with just an inert H-atom for a side "chain," will just be a "random coil" of protein that changes shape a lot. Think of a rope or a snake.
But side chains that can bond to each other can produce some really interesting structural elements. The dominant one's are the alpha helix (α-helix) and the beta-sheet (β-sheet).
2˚ structure: The α-helix

Image: National Institutes of Health/Wikipedia Commons
When the 1˚ structure will allow it, α-helices will form in a protein chain. A simplified version of an α-helix is shown on the left.
- Black = Cα
- Gray = Cβ
- Blue = amine nitrogen (N-H)
- Purple = amine hydrogen (N-H)
- Red = oxygen (O)
- Green = side chain (for simplicity here all side chains are green spheres)
An α-helix forms when the side chains are of the type that will tend to interact favorably with the water in solution. Notice that all of the side chains in this helix are pointing away from the helix. That occurs when side chains are mostly polar are charged (acidic or basic).
When that's the case, the amine proton (H) on every fourth AA is able to form a H-bond with the carbonyl oxygen (O) of the AA four places along. Take a minute to stare at the figure to see the repeated sequence: N-H --- O - Cβ - Cα - N-H --- and so on, where the (---) denotes a hydrogen bond.
If the side chains can orient outward toward the solution, this accident of the geometry of protein chains makes α-helices easy to form.
But if there are a large number of aliphatic or aromatic side chains present in the sequence, these tend to orient themselves inward toward the middle of the protein, not outward toward the solid. It's a case of "like dissolves like," or the hydrophobic effect.
If enough amino acids orient inward, an a-helix will not form, so the α-helix form of secondary structure is dependent on the primary structure.
Below are some examples of sequences that do and do not form α-helices.

The top three short protein sequences above are known to form α-helices. The blue residues are acidic or basic, and the yellow ones are charged. These residues are either charged or polar, so they will orient toward the water solvent, allowing an α-helix to form. There aren't enough aliphatic residues (not colored) in these sequences to break up helix formation.
The bottom sequence contains a few tyrosines, which contain an OH group and are somewhat soluble in water, but those are insufficient to overcome the propensity of the many aliphatic amino acids to bury themselves inside the protein and break up any helix formation.
While it is possible to predict where helices will form in a protein based on primary structure, that kind of prediction is still not that accurate. If we want to know the structure of a protein, we stil have to measure it using a technique like x-ray crystallography, nuclear magnetic resonance (NMR) or high-resolution electron microscopy (EM).
Proline is a "helix buster"
Proline is unique among amino acids because its amine group is tied down and cannot H-bond the same way as other amino acids in an α-helix. For this reason, α-helices never contain prolines. In fact, a proline can interrupt a helix very briefly, forming a kink in it.

Terminology: N & C-termini
Because of the way peptide bonds form, one end of the protein always ends in an H2N- group, the other in a -COOH. These are called the N-terminus and C-terminus, respectively, and we usually write proteins in the N→C direction.
2˚ structure: β-sheets

β-sheets or "β-pleated sheets" are another important motif of secondary structure in proteins. In certain conditions, again depending on the identity of side chains, strands of a protein will line up, either parallel (both N-termini on the same end) or antiparallel (N→C to C→N).
It is difficult to predict where β-sheets will form in a protein chain just by looking at the sequence. Sometimes, sheets are formed from far-separated parts of the chain, with a loop of relatively unstructured protein, or perhaps an α-helix, intervening between strands (β-strands) of the sheet.
There is a wide variety of β-sheet motifs across many proteins. They often impart the majority of the central structure of a protien, with other structural elements protruding from them. Here are just a few examples, illustrated as ribbon diagrams. In a ribbon diagram, all identity of the residues of the protein chain is lost in order to give a clear picture of the basic structure of the protein backbone.

3. Tertiary (3˚) structure
The tertiary structure (3˚ structure) of a protein describes the ways that various elements of secondary structure, α-helices, β-sheets and other nonstructured regions, interact with one another to form a completely "folded" protein.
These interactions depend, once again, on the locations and properties of the side chains, particularly polar and charged (acidic or basic) side chains that are more likely to form mutual attractions.
The tertiary structure can included things like bundles of α-helices held together by inter-helix bonds between side chains, helices that closely associate with β-sheet elements, regions of relatively unstructured rope-like protein chain sometimes referred to as "random coil", and even protein and its 2˚ structure elements that bind to and orient some molecule like the porphyrin hemoglobin that, in turn, binds to oxygen inside red blood cells.
Here are several examples of proteins, shown as ribbon diagrams for simplicity.
NF·κB (NF-kappa-B, or nuclear factor κB)→, is a DNA-binding protein that works inside the nucleus of cells (but doesn't reside there all the time). It's shown here bound to a short section of DNA using small loops of protein that form charge-charge and H-bond interactions with the bases and phosphate backbone of the DNA. The purple and green molecules in this version of the protein are identical; two NFκB monomers bind to DNA as a dimer.
Notice the many β-strands that form what's known as a globular protein. The 2˚-structure β-strands are pulled into this configuration by interactions between the side chains that stick out from them. Likewise, the two α-helices near the bottom of each monomer are held in place by those kinds of interactions.
The tertiary structure of proteins is what gives them their unique 3-D structure, and thus their function, but that structure is, of course, completely dependent on the sequence of amino acids in the chain.


← Aquaporins are a class of proteins that look like this one, made of bundles of α-helices that bury themselves in a cell membrane and form a channel for water molecules to pass selectively through.
Inter-helix attractions between side chains hold these helices into the fairly rigid configuration shown here.
4. Quaternary structure
The aquaporin protein above is actually formed from several protein monomers; this one has six. That's actually a fourth level of protein structure, tertiary (4˚) structure, the sticking together of more than one complete protein to form a complex of proteins.
Hemoglobin ( → ) is the carrier of oxygen (O2) in blood, transporting it from the lungs (or gills) to the tissues that need it.
Hemoglobin is a ternary (contains four subunits) complex of two pairs of similar molecules called the α and β subunits (red & blue, respectively). The 2˚ structure of each subunit, mostly α-helices, is bound into a globular protein by inter-helix side chain interactions to form the 3˚ structure of each. Each is then attracted to two others and binds in a similar manner to form the 4˚ structure you see.
The green parts of this structure are heme molecules, non-protein molecules bound to the protein to help it do its job. Many proteins bind other non-protein elements, called ligands, from single atoms to larger molecules. In this case the heme is what carries and releases the O2 molecules carried by hemoglobin.

Summary: Levels of protein structure
| Primary | 1˚ | The sequence of amino acids in the chain |
| Secondary | 2˚ | α-helices and β-sheets form where favorable conditions exist |
| Tertiary | 3˚ | Side chain interactions create a characteristic 3D structure by binding 2˚-structure elements together, burying hydrophobic residues in the core and exposing hydrophillic residues to the solvent |
| Quaternary | 4˚ | Individual proteins, and perhaps metal atoms and/or non-protein molecules (ligands) bind together in a complex |
xaktly.com by Dr. Jeff Cruzan is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. © 2016, Jeff Cruzan. All text and images on this website not specifically attributed to another source were created by me and I reserve all rights as to their use. Any opinions expressed on this website are entirely mine, and do not necessarily reflect the views of any of my employers. Please feel free to send any questions or comments to jeff.cruzan@verizon.net.
